Resarchers at University of Adelaide in Australia and Stanford University in California developed a more efficient process for producing genetically designed bacteria. The team led by Adelaide biotechnology professor Keith Shearwin published its findings online earlier this month in the journal ACS Synthetic Biology (paid subscription required).
Shearwin and colleagues call their process “clonetegration,” which simplifies the cloning of DNA, by integrating one or more additional DNA fragments into the genomes of bacteria. The team says its process makes possible the cloning and expression of genetic components usually difficult to propagate in plasmids, the strands of DNA occurring independently of chromosomes in bacterial genomes.
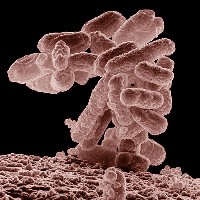
In the paper, the researchers demonstrate their process on Escherichia coli (E. coli), popularly known as an infectious bacteria in food, but also a common test model in biology labs. Shearwin says this process can be applied to the development of therapeutic biological compounds, such as insulin.
Among the advantages of the new process, says Shearwin, is its speed. He notes current gene-integration techniques require several days, while the team’s methods can be run overnight. In addition, the Adelaide-Stanford process allows for multiple rounds of DNA integration in the same bacteria, as well as simultaneously integrating multiple genes at different locations in the genome.
“This will become a valuable technique for facilitating genetic engineering,” says Shearwin, “with sequences that are difficult to clone as well as enable the rapid construction of synthetic biological systems.” A University of Adelaide statement says the molecular tools involved in clonetegration will be made freely available for other researchers and further development.
Read more:
- Synthetic Biology Census Shows Company Growth, Consolidation
- Oregon Health, Intel Partner on Genome Analysis Computing
- Statistical Database Analysis Links Genes, High Cholesterol
- Modeling, Biotechnology Boost Antibiotic Impact on Pathogens
- Study to Genetically Alter Algae for Faster Biofuel Output
* * *

 RSS - Posts
RSS - Posts
You must be logged in to post a comment.