29 December 2015. A bioinformatics team at St. Jude Children’s Research Hospital created a Web-based system that illustrates genetic mutations behind pediatric cancers. The software and database in ProteinPaint, as the system is called, are described in a letter in today’s issue of the journal Nature Genetics (paid subscription required).
The team led by Jinghui Zhang, chair of St Jude’s computational biology department, developed ProteinPaint to make better sense of the large-scale and complex genomic data collected about cancer and human disease overall. The system, made freely available to academic researchers, charts mutations of individual genes that alter their instructions for encoding proteins. St. Jude is seeking partners for commercial licensing and marketing of ProteinPaint.
“We developed ProteinPaint as an intuitive tool any scientist can easily use to explore the vast amount of information now available on cancer genomics,” says Zhang in a St. Jude statement. “Novel tools are essential to help scientists use this wealth of genomic data to advance research and find new cures.”
ProteinPaint is based on data from Pediatric Cancer Genome Project, a joint effort with Washington University in St. Louis, and National Cancer Institute’s TARGET database, short for Therapeutically Applicable Research to Generate Effective Treatments. Pediatric Cancer Genome Project collects genomic data on cancerous and healthy cells from more than 800 children. TARGET collects genomic data on childhood cancers from a network of participating researchers. ProteinPaint also stores data from other published studies.
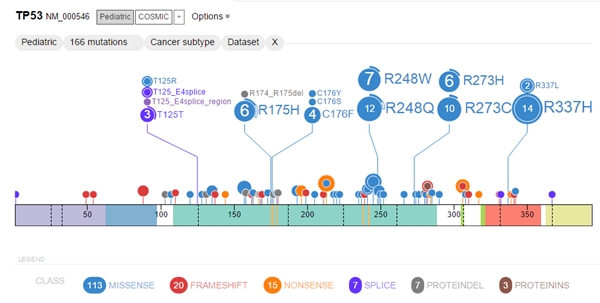
The database now stores data on some 27,500 mutations from more than 1,000 patients with 21 different types or sub-types of cancer. The database also stores RNA-sequencing data from 928 pediatric tumors in 36 cancer sub-types, which show how instructions encoded in DNA are transcribed into RNA molecules that send instructions to cells. The system integrates these data, then allows researchers to query the database to learn more about specific mutations.
Results are displayed with graphical tools that the researchers say are not available in current visualization programs.Details provided include mutation type, frequency in cancer sub-type and location in the protein domain. Data from ProteinPaint can show, for example, if mutations occur throughout the genome or only in cancer cells, as well as the role of mutations in the cancer’s beginning, progression, or relapse.
Researchers from St. Jude and Washington University already used ProteinPaint to reveal new insights into the extent of cancer-causing mutations, suggesting more children may be susceptible to cancer than previously thought. In a recent New England Journal of Medicine article, the team reports 8.5 percent of patients show mutations of genes in their normal tissue that increase their risk of developing cancer. In addition, more than half of the children with these systemic mutations lack a family history of cancer. Up to now, these mutations were considered to be extremely rare or occurred in families with a history of cancer.
Read more:
- Advocacy Group, Genetics Company Partner on Brain Disorder
- Genomic, Patient Data Integrated for Precision Medicine
- Spin-Off to Provide Lower-Cost Molecular Imaging Technology
- Full Genome Sequencing Offered for Under $1,000
- Genomic Sequencing Improves Children’s Cancer Care
* * *


 RSS - Posts
RSS - Posts
You must be logged in to post a comment.