Subscribe for email alerts
Donate to Science & Enterprise
|
By Alan, on December 11th, 2023  (Skeeze, Pixabay) 11 Dec. 2023. I started this publication, then called Science Business, in July 2010 to report each day on a fascinating topic, and in an honest and straightforward way. After 13 and a half years, the intersection of science and business remains a compelling subject, but there are other things I now want to do, and it’s time to bring Science & Enterprise to a close.
Science & Enterprise tells stories of people worldwide solving difficult problems with genius and hard work, but also about their support system in finance and government. The economic engine of advanced societies in the 21st century may be the brainpower of their people, but history shows turning good ideas into solutions requires a marketplace. The free market provides an efficient way for people with good ideas to connect with investors willing to back those ideas with real money, to create products and services with real value.
Good ideas from science often begin in university labs, where researchers seek to create new knowledge as well as attack specific problems. Where those ideas may have market value, top research universities in the U.S. and elsewhere now encourage researchers to become entrepreneurs and start companies to commercialize their findings, which make up many of the stories published here. And Science & Enterprise reports on financial and regulatory developments that affect businesses advancing science to the marketplace.
Respect the reader’s time and intelligence
I first became aware of these research entrepreneurs in my previous work as managing editor of Science Careers, the careers section of Science magazine, where starting a company provides an alternative career option to academic science. And before then, I wrote regularly on information technologies for a business audience. But in this publication, I also applied a honest definition of news that respects the reader’s intelligence. Plus, I tried to respect the busy reader’s time, thus stories are usually kept to about 500 words.
In Science & Enterprise, you find reporting six days a week on tangible advances in science or a business, without speculation or hype. Stories cover research by company scientists in academic journals or scientific meetings, new business formations, collaborations between business and academic researchers, patents awarded, licensing deals, early venture financing, IPOs, mergers, and regulatory actions.
Because most research, at least in the U.S., is health-related, so are most of the stories in Science & Enterprise. Whenever possible, I looked for more innovative solutions rather than more of the same, and the earlier the better. As a result, this publication first reported on a start-up biotech called Moderna and its messenger RNA therapeutics in 2012 and Crispr genome editing in 2013. And in Jan. 2020, I reported on a “novel coronavirus” emerging in the previous month from Wuhan, China, causing a mysterious disease that became known as Covid-19. Science & Enterprise also reported on new financing mechanisms, particularly challenge competitions that promote faster development of scientific breakthroughs.
What’s next? I’ll probably pursue my photography interests, but with more emphasis on visual storytelling and publishing than just snapping photos. Whatever happens will occur under Technology News and Literature, the publisher of Science & Enterprise.
Let me thank the Science & Enterprise readers, particularly subscribers through WordPress or email, as well as advertisers and agencies over the years. And a special thanks to my wife Sharon, who had to listen to my recitation of daily stories in this publication for 13 and a half years. I’m sure we’ll find much more to talk about over dinner.
* * *
By Alan, on June 12th, 2024 – Sponsored content –
 (Gerd Altmann, Pixabay. https://pixabay.com/illustrations/digitization-particles-smartphone-7261158/) The recent explosion in new uses for artificial intelligence or A.I. has generated more attention to the models or algorithms that power these applications. Most of these new applications use a form of A.I. called machine learning, but their success depends on the soundness of the data that feed the models.
Machine learning, as the name implies, uses algorithms that learn, or adjust their outcomes as they encounter more and more data. As the algorithms process more data, their calculations should become more refined and precise. For example, some A.I. medical diagnostics analyze images captured from echocardiograms that display blood flow and heart mechanics in real time. For these algorithms, the wider the range of heart conditions and disorders shown in the images, the greater the scope of the algorithm and the likelihood of returning a true-positive or true-negative diagnosis.
Generative A.I. performs projections from the data used to train the algorithms, sometimes taking the form of text or images that meet user specifications. In these and many other machine learning models, raw-data inputs need context to maximize their usefulness to data scientists. Adding this context to raw data is a process called data labeling, considered a key step in training machine learning algorithms.
While some data labeling processes can be automated, human intervention is often needed to ensure the data that trains machine learning algorithms accurately reflect real-world conditions. Data labeling is nonetheless a process used occasionally by algorithm developers, and thus outsourced in many instances. As a result, choosing a data labeling service requires model designers to look into the organizational structure and management of their outsourcing partners, much like they would hire their own professional staff.
When reviewing data labeling services, consider factors such as relationship of labelers to the company — direct-hires or crowd-sourced contractors — and presence of on-site management. In addition, security and integrity of data in the hands of the outsourcing partner need to be guaranteed. Visit the web site of Innovatiana for an example of a data labeling company that meets these specifications.
* * *
By Alan, on May 28th, 2024 – Sponsored content –
 (Rawpixel, Unsplash) 28 May 2024. With the globalization of trade, businesses are increasingly looking beyond their national borders to explore new markets, tap into diverse consumer bases, and seek growth opportunities. However, these opportunities come with inherent risks. International business risk refers to the uncertainties and potential adverse consequences that organizations face when operating across borders. These risks may arise from a variety of sources, including economic, political, cultural and operational factors. Below we will look at the main factors and classifications of risks associated with conducting business activities on the world stage.
Our company is a team of specialized specialists offering a wide range of corporate services. We are dedicated to achieving the most effective and optimized results for you.
International business risks
One of the common international business challenges that many companies face is political and economic instability in various regions of the world. Political and economic instability may affect the business environment, market demand, foreign exchange rate, legal and regulatory frameworks, employee and asset security, and the overall profitability and sustainability of business operations. It is therefore important for international businesses to have a clear understanding of the political and economic risks and opportunities in each country in which they operate, and to develop effective strategies to mitigate negative impacts and exploit positive ones.
Entering a foreign market initially implies competition with local companies. Understanding the competitive environment is very important for finding potential partners or collaborators, as well as for developing effective marketing strategies. To be successful, foreign companies must also have a sufficient understanding of customer preferences. Conducting market research can help companies identify potential opportunities and develop effective marketing strategies.
Also important and carrying a special meaning is the risk of changes in exchange rates. Entering the international market means working with different currencies. Any change in exchange rates can affect a company’s profits and lead to losses. A risk management strategy must be developed to minimize potential losses.
Insufficient staff training is also a risk. International business requires employees to have specialized knowledge and skills. Staff need to be trained to ensure they are ready to work internationally.
And another important risk aspect is political and economic instability. There may be political and economic crises in different countries that can affect business. It is necessary to monitor the political and economic situation in the countries where the company operates and develop action plans in the event of a crisis.
Hedging strategies to mitigate foreign exchange risks and how to manage economic risk from tax changes
Managing and mitigating risks in these categories may involve implementing the following strategies and approaches.
- Diversification of business portfolio and income streams across different regions and markets. This can help reduce dependency and exposure to one or more markets that may be affected by political and economic instability, and increase business resilience and agility to adapt to changing conditions and opportunities. Diversification can also help balance the risks and rewards of different markets and sectors, and optimize the allocation of resources and capital.
- Setting clear budget boundaries. Always make a budget, regardless of your personal ambitions and desires. This applies to absolutely all aspects, from renting an office and finding specialists to join your team, to selling your own services or products. Don’t take it all on yourself; delegate some of the tasks to your sales manager. Of course, this approach will reduce the amount of expected profit, but on the other hand, it will reduce the level of responsibility; in any case, profit will be received daily. Before making global plans, look at how things are going in your company; if a product does not work in a specific foreign market, you will understand this long before the company’s large-scale withdrawal. A SWOT analysis will not be superfluous; it will provide clear strategic planning and show the weaknesses of business development.
- Development of a business model for the market. Create a business model that suits the country and its demographics. Large countries with different market segments and geographies may require a multi-part model that includes specific strategies for each region. Assign a new product introduction team to determine the right business models, operating models, and profit expectations before entering the target market. The NPI team must tailor the product or service to meet the needs of the market and target customers.
- Emergency plan. When you decide to develop a business in a foreign market, make sure once again that you really need it. It is important to study the possible risks and benefits of the business at the initial stage. It’s good to be an optimist, but in business development and investment it is important to be realistic; believe me, even large companies have failures.
Finding a right partner and other ways of reducing risks in international business
Let us highlight several strategies that relate to issues not only of the financial side, but also of the partnership and legal side.
- Conducting a thorough assessment of political and economic risks before entering a new market or expanding an existing one. This can help identify potential threats and opportunities such as political stability, corruption, human rights, trade barriers, taxation, inflation, currency fluctuations, etc., and assess their likelihood and impact on business goals and operations.
- Developing a contingency plan for various scenarios and events that could disrupt business operations, such as civil unrest, violence, terrorism, natural disasters, sanctions, embargoes, etc. This can help prepare for worst-case scenarios and minimize losses and damages. The contingency plan should include alternative suppliers, distributors, customers, locations, modes of transportation, communication channels, etc.
- Building and maintaining strong relationships with local stakeholders such as government officials, regulators, business partners, customers, suppliers, employees, communities, media, etc. This can help build trust, authority, legitimacy and support business operations and facilitate access to information, resources and opportunities.
- It is necessary to develop a mechanism for positive interaction with employees, clients, suppliers and create standardized communication documents. It is important to stabilize the supply chain, as well as optimize the customer service management system in order to avoid the formation of a negative public opinion about the company.
- The physical and mental health of employees should be maintained. It is necessary to create a flexible work schedule and provide staff with all the technical means so that they can work remotely, if necessary.
- It is important to emphasize plans to respond to risks in supply chains. Multinational companies usually make provisions in advance for the use of redundant office space, production facilities and supply chains in different countries or regions.
- Before entering a new market, it is important to conduct thorough market research and analysis to identify potential opportunities, risks, and barriers that may impact the business. This includes understanding the local culture, consumer preferences, demand, competition, legal environment and trade policies.
Thus, risks must be assessed and mechanisms and plans for emergency response and work sharing must be determined. Many multinational companies have already developed a contingency plan or business sustainability plan and are prepared to implement it if necessary. If there is no such plan, then the company needs to immediately assess all risks, including problems related to personnel, outsourcing, relations with government agencies, public relations, and supply chain.
Risk assessment tools and systems
- SWOT analysis. One frequently used tool is a SWOT (Strengths, Weaknesses, Opportunities and Threats) analysis. It helps a business assess internal strengths and weaknesses as well as external opportunities and threats in the international market and develop strategies to mitigate risks.
- PESTEL analysis. Another valuable framework is PESTEL (political, economic, social, technological, environmental and legal) analysis. This tool examines external factors that can affect international business operations. It provides a comprehensive understanding of macro-environmental factors.
- Assessing the country risk associated with specific countries is important for international business. Factors taken into account include political stability, economic performance and cultural considerations.
Thus, we have identified potential risks for international business and possible ways to reduce them. The development and implementation of any strategies depends on the individual parameters of each business. Our experts will help you assess your project for potential risks and develop the best mitigation strategies for you. Contact us now.
Article’s author is Denys Chernyshov – founder and CEO of the globally-famous organization Eternity Law International.
* * *
By Alan, on April 15th, 2024 – Sponsored content –
 (Elchinator, Pixabay. https://pixabay.com/illustrations/periodic-table-chemistry-science-3962844/) 15 Apr. 2024. High-Performance Liquid Chromatography (HPLC) stands as a cornerstone of analytical chemistry. This powerful technique separates complex mixtures into their individual components, allowing researchers to identify and quantify them with remarkable precision. However, even the most sophisticated HPLC system is susceptible to wear and tear, leading to malfunctions that can significantly impact research progress. Understanding these common issues is crucial for maximizing uptime and ensuring the quality of data generated.
Where HPLC Reigns Supreme: Exploring Applications
HPLC plays a vital role in diverse scientific fields, including:
- Pharmaceutical Science: HPLC is indispensable for drug discovery and development. It assists in the purification, analysis, and quality control of drugs and their metabolites.
- Environmental Monitoring: HPLC is a valuable tool for identifying and quantifying pollutants in water, soil, and air samples. This information is crucial for environmental protection efforts.
- Food Science: HPLC helps ensure food safety by detecting contaminants, analyzing food composition, and monitoring food quality.
- Biotechnology: Researchers utilize HPLC to analyze proteins, peptides, and other biomolecules, aiding in fields like drug discovery and protein engineering.
- Forensics: HPLC can be used to identify drugs and toxins in biological samples, aiding in criminal investigations and forensic analysis.
The critical nature of HPLC in these diverse fields highlights the importance of timely troubleshooting and maintenance.
When the Flow Falters: Common HPLC Issues and their Impact
Several issues can disrupt the smooth operation of an HPLC system, potentially leading to inaccurate results and research delays. Here’s a closer look at some common problems:
- Peak Broadening: Broadened peaks indicate a loss of resolution, making it difficult to distinguish between closely eluting components. This can be caused by:
- Clogged frits: The frit is a small filter at the inlet and outlet of the column. Clogging can restrict flow and reduce column efficiency.
- Degraded stationary phase: The stationary phase is the “heart” of the column, where separation occurs. Over time, the stationary phase can degrade, leading to peak broadening.
- Incorrect mobile phase composition: The mobile phase is the solvent that carries the sample through the column. Choosing an inappropriate mobile phase composition can affect peak shape and resolution.
- Ghost Peaks: These unwanted peaks appearing on the chromatogram indicate contamination in the system. Sources of ghost peaks include:
- Dirty injector needle: Sample residue on the injector needle can carry over to subsequent injections, causing ghost peaks.
- Carryover from previous samples: Incomplete cleaning of the system after injecting a concentrated sample can lead to carryover and ghost peaks in subsequent injections.
- Contaminated mobile phase: Impurities in the mobile phase can also contribute to ghost peaks.
- Detector Issues: Depending on the type of detector used (UV/Vis, refractive index, etc.), various malfunctions can occur:
- Lamp issues: UV/Vis detectors rely on a light source (lamp) to detect analytes. A malfunctioning lamp can lead to reduced sensitivity or erratic baseline noise.
- Detector electronics: Electronic malfunctions within the detector can affect signal processing and data acquisition.
- Misalignment: Misalignment of detector components can impact sensitivity and lead to inaccurate results.
Beyond the Peak: Additional Concerns and Troubleshooting Tips
While peak broadening, ghost peaks, and detector issues are prominent concerns, other potential problems require attention:
- High System Pressure: Excessive pressure can damage the column and reduce separation efficiency. Causes include clogged frits, a packed column, or an inappropriate mobile phase viscosity.
- Leaks: Leaks in the system can disrupt flow, reduce accuracy, and waste mobile phase. Regularly checking for leaks at connections and fittings is crucial.
- Retention Time Shifts: Retention times indicate how long it takes for a component to elute from the column. Unexpected shifts can be caused by changes in column temperature, mobile phase composition, or flow rate.
Troubleshooting Tips:
- Consult the Manual: The manufacturer’s manual is a valuable resource for troubleshooting common issues and proper maintenance procedures.
- Maintain a Maintenance Log: Documenting maintenance activities, observations, and any troubleshooting steps taken aids in identifying recurring problems and scheduling preventive maintenance.
- Invest in Training: Proper user training on operating procedures and basic troubleshooting techniques can help prevent user errors and identify potential problems early on.
The Preventive Approach: Maximizing Uptime and Data Integrity
While addressing specific issues is crucial, a proactive maintenance approach with a provider such as Peak BioServices (https://peakbioservices.com/hplc-repair-service-maintenance/) is essential for maximizing HPLC system performance and data integrity. Here are key strategies:
- Regular Cleaning: Routine cleaning of the injector, detector flow cell, and column is vital to prevent contamination and ensure optimal performance.
- Mobile Phase Filtration: Using high-quality filters for the mobile phase helps remove contaminants that can contribute to peak broadening or ghost peaks.
- Column Care: Proper column care is essential for long lifespan and optimal performance. This includes following recommended storage conditions, avoiding excessive pressure, and using appropriate mobile phase compositions.
- Regular Calibration: Regular calibration of pumps, detectors, and temperature sensors ensures the accuracy of measurements and data generated by the HPLC system.
- Preventive Maintenance: Investing in preventive maintenance by qualified service engineers allows for comprehensive inspections to identify potential problems before they escalate into major breakdowns. This can include checking for leaks, verifying flow rates, and evaluating column performance.
A Symphony of Precision: The Importance of a Reliable HPLC System
HPLC plays a critical role in various scientific endeavors. Its ability to separate complex mixtures with remarkable precision makes it an indispensable tool for research and analysis. However, even the most sophisticated system requires ongoing care and maintenance. By understanding common issues, implementing a proactive maintenance strategy, and prioritizing user training, researchers can ensure the smooth operation of their HPLC system. This, in turn, guarantees the generation of accurate and reliable data, ultimately contributing to advancements in various scientific fields.
The analogy of an orchestra aptly describes the role of HPLC in research. Just as each instrument plays a vital role in producing harmonious music, a well-maintained HPLC system contributes to the symphony of scientific discovery. By ensuring its optimal performance, researchers can focus on their experiments, confident that their HPLC system is a reliable partner in their scientific journey.
* * *
By Alan, on February 29th, 2024 – Sponsored content –
 (Julita, Pixabay. https://pixabay.com/photos/park-bike-senior-lonely-cycling-5528190/) 29 Feb 2024. According to recent data, the number of courses dedicated to the over-65s and those aimed at weight loss is on the rise. Still, awareness of the importance of exercise for psychological well-being is growing.
Besides, more and more seniors are also looking for natural components, and CBD oils are now used for muscle relaxation, better sleep, and more! The only important thing is to choose the right supplier from several CBD Flower Shops!
Stay healthy!
The ‘recipe’ for longevity also passes through regular and constant physical activity, and a survey in the scientific journal ‘Health and Fitness Journal’ indicates the following trends in health and fitness. Although it is an essential tool, especially for wellness centers and fitness professionals, the report also helps citizens orient themselves among the new trends, especially at the beginning of the year when one good intention is to join a gym or a sports course. Here, then, are the leading fitness trends worldwide in 2024 and their respective health benefits.
Wearable technology devices
The use of smartwatches, fitness trackers, and heart rate monitors continues to be a major trend, and now, in many of the larger fitness centers, cardio, and weight room equipment can be synchronized with smartphones and smartwatches to provide a fully connected workout experience and to track physical activity.
The various wearable devices may be fine for healthy people who do not have any particular health problems, but it is good to understand that everyone needs their personalized workout this cannot be done with apps and smartphones, but it is better to be followed by a motor science expert.
Please beware of the risk of bigorexia, a psychological disorder characterized by an obsession with one’s physical appearance and a constant preoccupation with appearing unmuscular or fit. It is a real addiction that can only be combated if families manage to recognize it early on.
Corporate wellness also in the UK
The trend also concerns the UK, as shown by the latest Report Benefits and Trends data, a survey conducted by Aon, an insurance brokerage, and risk management consultancy company, on a sample of 5,000 companies representative of the business fabric. According to the survey results, 79% of the organizations confirm that they have a Welfare strategy or intend to implement it. Among the most popular services to improve the work-life balance is the company gym or the possibility of having in-house wellness activities (10%).
Fitness programs for the over 70s
The new demographic picture is also reflected in the population’s general health because aging increases the risk of chronic diseases, cognitive impairment, and falls. ‘Doing physical activity is important at all ages, but even more so after the age of 60, and it is important to know what you need to exercise,’ Beltrami adds.
The workouts seniors need
Not just walking or aerobic activity, strength training is needed to avoid sarcopenia, i.e., the loss of muscle mass that causes a drop in metabolism, the onset of osteoporosis, and an increased risk of falling.
Are there any workouts that are more suitable than others for those over 65 years of age? Physical activity must be as individualized as possible because these individuals often already suffer from specific pathologies,’ replies the Fmsi vice-president. This is why a visit to the sports doctor, who can determine the most suitable type of training, is necessary. However, training muscular plasticity, aerobic capacity, and balance is essential.
Training for weight loss
The trends of 2024 could not fail to include the whole strand of fitness activities aimed at weight loss, which are the ones that make gym memberships soar after the holiday season. Sport and regular exercise help improve metabolism and fight fat!
To lose weight, diet alone often doesn’t work, and you need to combine it with exercise to increase muscle mass, which is directly proportional to calorie consumption; in fact, athletes can afford to consume a lot of calories because they have good muscle mass.
The medical prescription for physical activity
Among the trends are those relating to promoting physical activity among young people and the figure of personal trainers as health and fitness professionals with an accredited certification that can represent a guarantee of the quality of the services provided, given that health and wellbeing are involved. However, there is increasing recognition of the need to consider physical activity as a natural drug to be prescribed.
Where do we stand?
Although an active lifestyle is one of the most effective preventive treatments for chronic diseases, more than 25 % of adults must comply with current exercise guidelines; the Medical and Sports Federation has already drawn up guidelines prescribing physical activity for specific pathologies. We are tackling this path very carefully, involving sports physicians and exercise science graduates.
It would be essential to extend the first physical activity ‘prescription’ projects to a national level; we would need more funds because prevention is fundamental at all levels.
Exercises for mental health
The effects regular physical activity can have on psychological well-being is also a recent survey conducted by an association representing fitness operators, from which representing fitness professionals, which showed that more than half of people who join a gym do so to cope with illness, 78 % said that going to the gym has a positive impact on mental health. In comparison, two-thirds (66 %) said exercising helps them sleep better. Among the activities that most promote mental health are yoga, mindfulness, dance, and group classes.
And why not use JustBob.shop CBD oil to facilitate a better mood and good mental health in general.
* * *
By Alan, on December 8th, 2023  Killer T-cells surround a cancer cell (NICHD, Flickr) 8 Dec. 2023. The developer of an experimental immunotherapy delivering nanoscale payloads to treat pancreatic cancer says the treatment received fast-track status from the Food and Drug Administration. The biotechnology company EnGeneIC in Sydney, Australia is developer of cancer therapies delivered directly to tumor cells, guided by synthetic antibodies that it says avoid toxicity to healthy tissue.
The EnGeneIC technology packages cancer-killing treatments, either conventional chemotherapies or synthetic interfering RNA, as glycolipid or carbohydrate/natural oil particles about 400 nanometers across; one nanometer equals one billionth of a meter. The company says its EnGeneIC dream vectors or EDVs can concentrate up to one million molecules of the cancer-killing drugs, much higher concentrations than given systemically to patients, making them more potent in destroying tumors. EnGeneIC says at 400 nanometers, EDVs are too large to pass through blood vessel linings in healthy tissue, but will escape through in leaky blood vessels in microenvironments, the supportive mass of cells and blood vessels surrounding tumors.
The company says its EDVs are guided to tumors by synthetic antibodies designed to target epidermal growth factor receptor proteins, overexpressed on cell surfaces in the vast majority of solid tumors. Once on the cell surface, says EnGeneIC, the nanoscale therapy payloads are ingested through the cell membrane, then broken-down inside the cell, releasing their cancer-killing contents, thus minimizing toxicity elsewhere. In addition, says the company, the presence of EDVs creates a microenvironment more receptive to immune responses, with a preclinical study showing a greater production of dendritic, macrophage, natural killer, and tumor-specific T-cells that can prolong remission times.
EnGeneIC is testing its EDVs in a clinical trial among patients with advanced pancreatic ductal adenocarcinoma, the most common form of pancreatic cancer that begins in the lining of the ducts carrying enzymes from the pancreas. Pancreatic cancer in many cases is difficult to diagnose in its early stages, making treatments more problematic once detected in later stages. The early- and mid-stage trial is enrolling up to 40 patients in Australia with advanced pancreatic ductal adenocarcinoma or PDAC expressing epidermal growth factor receptor proteins. The study team is looking for safety indicators, such as adverse effects from the treatments to determine safe dose levels, as well as anti-cancer activity, immune responses, and survival times. The trial has no control or comparison group.
Six months vs. two months average survival time
Initial results of the trial, published last month in the journal Clinical Cancer Research, report on 25 enrolled patients, of which seven patients dropped out due to the rapid advance of their disease, and one withdrew consent. The study team reports that before the withdrawal of these participants, 19 of the 25 reported mild to moderate adverse effects, which were resolved within hours, but showed no other safety-related concerns. Patients also exhibited minimal to no toxicities from the treatments.
Moreover, eight of the 17 patients completing the trial survived more than six months, compared to historical average survival times of two months. In addition, say the authors, 80 percent of patients receiving EDV treatments maintained their body weights or gained weight, compared to the severe weight loss seen in most pancreatic cancer patients.
FDA awards fast-track status to experimental drugs for serious unmet medical conditions exhibiting superior effectiveness, fewer adverse effects, reduced toxicities, or meeting key public health priorities. Treatments with fast-track status gain more frequent meetings and written communications with FDA staff, and are eligible for expedited or rolling (stage-by-stage) reviews.
In an EnGeneIC statement released through Globe Newswire, the company says fast-track status from FDA will help expedite clinical trials of EDVs for pancreatic cancer patients in the U.S. Co-founder and CEO Himanshu Brahmbhatt notes, “We are committed to expeditiously advancing our clinical program for PDAC patients and others with low survival cancers. These are the patients who need it most.”
More from Science & Enterprise:
We designed Science & Enterprise for busy readers including investors, researchers, entrepreneurs, and students. Except for a narrow cookies and privacy strip for first-time visitors, we have no pop-ups blocking the entire page, nor distracting animated GIF graphics. If you want to subscribe for daily email alerts, you can do that here, or find the link in the upper left-hand corner of the desktop page. The site is free, with no paywall. But, of course, donations are gratefully accepted.
* * *
By Alan, on December 7th, 2023  (NIH.gov) 7 Dec. 2023. A developer of treatments for brain disorders and researchers at Stanford University aim to harness ultrasound to deliver drugs in nanoscale particles to precise regions of the brain. The collaboration brings together Cordance Medical in Mountain View, California with radiologist Raag Airan and neuroscientst Nolan Williams, members of the Stanford Medicine faculty.
Cordance Medical is a five year-old biotechnology company creating non-invasive devices for diagnosing and treating brain diseases. The company’s technology uses ultrasound to help drug treatments cross the blood-brain barrier, a collection of tightly packed cells lining blood vessels in the brain preventing foreign substances from entering the brain. Those foreign substances include most drugs, making it more difficult to treat brain disorders, including cancer and neurodegenerative diseases such as dementia and Parkinson’s disease. The blood-brain barrier also limits cells from brain tumors from entering the general blood stream, preventing the growing use of liquid biopsy blood tests to detect brain cancers.
Cordance Medical devices use ultrasound, a form of targeted sound waves, focused on precise areas of the brain to gently stretch blood vessels in those regions, allowing drugs to pass in or circulating tumor cells to pass out. Low-frequency ultrasound waves, says the company, create micro-scale bubbles that vibrate in the blood stream expanding blood vessels to allow drug molecules or circulating tumor cells to pass through. Patients wear a Cordance helmet-like headgear that concentrates the ultrasound waves on regions of the brain previously identified through MRI or CT scans. In October 2023, the company’s NeuroAccess device to enable liquid biopsies received a Breakthrough Device designation from the Food and Drug Administration.
Delivery with ultrasound to cerebrospinal fluid
In the collaboration with Stanford medical school, Cordance Medical is joining with researchers Raag Airan and Nolan Williams to study targeted drug delivery with nanoscale drug particles activated by ultrasound. Airan’s lab investigates precise drug delivery to the brain with both drug nanoparticles and ultrasound, with research published last year demonstrating the group’s techniques in lab rats delivering drugs to cerebrospinal fluid with ultrasound. Williams is part of Stanford’s Brain Stimulation Lab that studies neurostimulation as a psycho-pharmalogical process for treating neurological and psychiatric disorders.
The collaboration with the Stanford neuroscientists seeks to adapt Cordance Medical’s NeuroAcess device for releasing drug-loaded nanoparticles with ultrasound in precisely targeted brain locations. The company says this “uncaging” capability goes beyond the technology’s original blood-brain barrier objectives, to treat a range of neurological and psychiatric disorders, while minimizing adverse effects.
Ryan Dittamore, CEO of Cordance Medical, says in a company statement that the agreement is “a critical step forward in the field of precision neuropsychiatry.” Dittamore adds, “Our NeuroAccess platform, when paired with Dr. Airan’s novel nanoparticles, has the potential to revolutionize patient care by providing more precise control of neural activity.” The agreement, says the company, includes a first-in-human clinical trial in patients with chronic pain, testing a nanoparticle formulation of the drug ketamine.
More from Science & Enterprise:
We designed Science & Enterprise for busy readers including investors, researchers, entrepreneurs, and students. Except for a narrow cookies and privacy strip for first-time visitors, we have no pop-ups blocking the entire page, nor distracting animated GIF graphics. If you want to subscribe for daily email alerts, you can do that here, or find the link in the upper left-hand corner of the desktop page. The site is free, with no paywall. But, of course, donations are gratefully accepted.
* * *
By Alan, on December 6th, 2023  Natural killer cell (NIAID, Flickr. https://flic.kr/p/Lnh5i1) 6 Dec. 2023. A developer of treatments for cancer and autoimmune disorders from stem cells says FDA cleared its request to start a clinical trial of its therapy for systemic lupus erythematosus. Century Therapeutics, a biotechnology company in Philadelphia, says the Food and Drug Administration authorized a study of its experimental drug code-named CNTY-101, already in an early-stage trial for B-cell lymphoma, a blood-related cancer.
Systemic lupus erythematosus or SLE is the most common form of the chronic autoimmune disease lupus. An autoimmune disorder occurs when the immune system attacks healthy cells or tissue rather than an invading pathogen. In the case of SLE, connective tissue like joints or skin are affected, with symptoms like arthritis pain or skin rash, but also hair loss, sores, swollen glands, fatigue, and inflammation of linings of the heart or lungs. The disease can also lead to kidney damage, heart disease, and breathing difficulties. SLE is a complex disease, with no known single cause, but believed to be a combination of genetic and environmental triggers.
Century Therapeutics is a five year-old company spun-off from labs at Massachusetts General Hospital in Boston and Stanford University in Palo Alto, California. Immunologist Marcela Maus at Mass. General and Harvard Medical School, one of the company’s scientific founders, studies the biology and engineering of T-cells in the immune system as cancer therapies, while cell biologist Hiromitsu Nakauchi at Stanford medical school, also a scientific founder, investigates clinical applications of stem cells. Century Therapeutics licenses technology for stem cell transformation and production from Fujifilm Cellular Dynamics in Tokyo. Science & Enterprise reported in July 2019 on the company’s raising of $250 million in its first venture round.
Gene edits to evade immune detection
CNTY-101, Century Therapeutics’ lead product, is made from off-the-shelf induced pluripotent stem cells, also known as adult stem cells, transformed into natural killer cells in the immune system. The company says it uses the genomic editing technique Crispr, short for clustered regularly interspaced short palindromic repeats, to edit the genes in stem cells to target their therapeutic effects, as well as evade detection from the immune system called Allo-Evasion, allowing the therapies to work. CNTY-101 is being tested in an early-stage clinical trial at two sites among patients with B-cell non-Hodgkin’s lymphoma, begun earlier this year, looking mainly for safety and tolerated dosage, but also clinical responses to the treatments.
Century Therapeutics says FDA cleared its investigational new drug or IND application for a similar early-stage clinical trial testing CNTY-101 among patients with SLE. The new trial, says the company, will investigate the treatment’s safety and tolerability in participants, as well as chemical activity in the body and any clinical responses. Century Therapeutics expects the study to begin in the first half of 2024, with initial results by the end of the year.
“We believe the unique profile of CNTY-101,” says Century Therapeutics CEO Brent Pfeiffenberger in a company statement, “which incorporates multiple precision edits including our Allo-Evasion technology, positions it as an off-the-shelf allogeneic treatment option that could meaningfully improve outcomes for patients with SLE for whom existing therapies fall short.”
More from Science & Enterprise:
We designed Science & Enterprise for busy readers including investors, researchers, entrepreneurs, and students. Except for a narrow cookies and privacy strip for first-time visitors, we have no pop-ups blocking the entire page, nor distracting animated GIF graphics. If you want to subscribe for daily email alerts, you can do that here, or find the link in the upper left-hand corner of the desktop page. The site is free, with no paywall. But, of course, donations are gratefully accepted.
* * *
By Alan, on December 4th, 2023 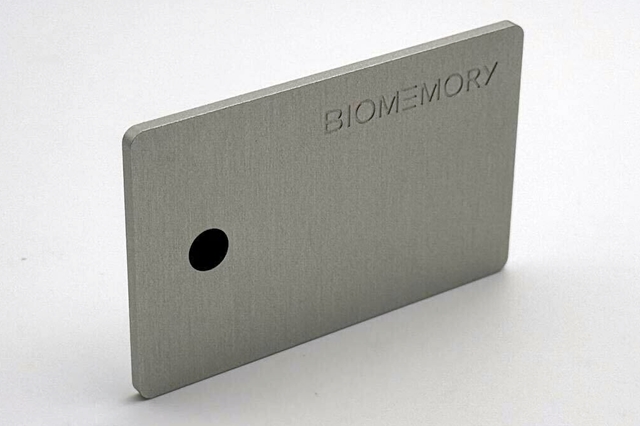 DNA Card (Biomemory, Cision) 4 Dec. 2023. A company developing DNA into a feasible method for large-scale data storage unveiled a credit card-sized device using DNA for off-line data storage. Biomemory, a two year-old biotechnology company in Paris, now offers its DNA Card that the company says can hold 1 kilobyte or 1,000 characters of text, as a practical demonstration of its technology.
The company says DNA data storage makes it possible to store digital data at one million times more density than current media, such as solid-state drives or magnetic tape, to meet a rapidly growing need for reliable data storage. And, says Biomemory, DNA offers a much more durable medium for data storage than current technologies, as evidenced by the ability to retrieve and decode DNA from fossils of extinct animals unearthed after many thousands of years. In addition, the company says its process is environmentally friendly and remains stable at ambient temperatures, without an external energy source, and emits no carbon dioxide.
Science & Enterprise reported on Biomemory in Nov. 2022, when the company raised €5 million in seed funds. At the time, researchers from Biomemory had recently posted a manuscript describing the company’s process called DNA Drive that uses an algorithm to encode binary data into DNA sequences. Biomemory creates synthetic DNA reflecting those sequences for assembly into bacterial DNA plasmids, where the encoded DNA sequences are amplified and extracted for storage. When needed, the company’s algorithm decodes the stored DNA sequences back into binary data.
DNA extracted and decoded
The DNA Card, says Biomemory, uses this process to create a chip in the card that stores data specified by the user in synthetic DNA. Individuals send the company one kilobyte of data for encoding into DNA, equivalent to a short paragraph of text, that Biomemory returns to the individual encoded into DNA on two identical cards. The company then asks the recipient to send one of the cards to its partner Eurofins Genomics that provides genomic sequencing services. Eurofins Genomics extracts the DNA from the customer’s card, sequences the DNA, and returns the text to the user for comparison with the original. Biomemory charges a fee of €1,000 ($US 1,080) for the service.
While the amount of data on a DNA Card is limited, Biomemory says it’s still a major advance in DNA data storage. “After years of talk about the potential of molecular computing,” says Biomemory CEO Erfane Arwani in a company statement released through Cision, “we are incredibly proud to bring the first DNA data storage product to market, that not only pushes the boundaries of innovation but also aligns with our commitment to environmental sustainability and efficiency.”
The company says it’s scaling up the process to met the constantly expanding storage needs of data centers. By 2026, says Biomemory, the company plans to unveils its Biomemory Prime service for storing 100 petabytes of data. One petabyte is equivalent to 1,024 terabytes, with one terabyte equaling some 1 trillion bytes of data.
More from Science & Enterprise:
We designed Science & Enterprise for busy readers including investors, researchers, entrepreneurs, and students. Except for a narrow cookies and privacy strip for first-time visitors, we have no pop-ups blocking the entire page, nor distracting animated GIF graphics. If you want to subscribe for daily email alerts, you can do that here, or find the link in the upper left-hand corner of the desktop page. The site is free, with no paywall. But, of course, donations are gratefully accepted.
* * *
By Alan, on December 1st, 2023  (Manolo Franco, Pixabay) 1 Dec. 2023. A collaboration between biotechnology and dairy products businesses seeks to identify peptides in milk to form into dietary supplements for people with mood disorders. Financial and intellectual property details of the agreement between the biotechnology enterprise Lactocore Group and food products company Ingredia Dairy Experts SA were not disclosed.
Lactocore Group is a five year-old company in Boston and Berlin discovering therapeutic peptides, short chains of amino acids, for mental health and metabolic conditions in humans, farm animals, and pets. The company says it uses computational techniques to screen and identify peptides in milk with potential as therapies, followed by further behavioral screening with Zebrafish models and preclinical development in lab cultures and animals. The Lactocore technology is based on research in physiology, biochemistry, and molecular biology by the company’s founders at Moscow State University and elsewhere in Europe. In Aug. 2023, the company joined the Biolabs business accelerator in Heidelberg, Germany.
The company focuses on peptides because of their record of safety and tolerability, since they readily break down into amino acids that are easy to absorb. The lead product at Lactocore is a peptide code-named LCGA-17m16 to address anxiety and depression. The company says LCGA-17m16 limits the alpha 2 delta or a2d protein that acts on gamma-aminobutyric acid or GABA receptors in the brain. GABA receptors are nervous system activity inhibitors, kept in balance with excitatory proteins in healthy individuals. Several psychiatric disorders, including anxiety and depression, are associated with low GABA concentrations in the brain.
Extracts functional proteins and other bioactive ingredients
In Aug. 2021, Lactocore co-founder and CEO Anton Malyshev and colleagues in Russia and the U.S. reported in the journal Frontiers in Neuroscience on discovery and tests with lab mice of LCGA-17 peptide. The authors say the screening process shows LCGA-17 has a high affinity for a2d proteins and GABA receptors, and lab tests show the peptide binds with rodent brain cells comparable to leading psychiatric drugs. In further tests, lab mice induced with anxiety and depression, then given LCGA-17 peptides, are able to complete exercises indicating little anxious or depressive behavior.
The collaboration with Ingredia aims to develop food supplements enriched with therapeutic peptides to help relieve mood disorders in humans and animals. Ingredia, based in Arras in northern France, is a dairy cooperative that also extracts and markets ingredients from milk, including functional proteins and other bioactive ingredients. In their project, Lactocore and Ingredia are investigating peptides derived from cow milk after hydrolysis or chemical reactions in water. The team plans to screen the hydrolyzates with mass spectrometry, a process to identify the results at a fine level of granularity to isolate specific peptides for further development.
The project expects to begin in Jan. 2024. “Peptides have vast and diverse physiological effects in humans and animals,” says Malyshev in a Lactocore statement released through Cision. Malyshev adds, “This long-term partnership with Ingredia, an industry leader in the European segment of functional ingredients, marks a significant expansion in our research domain.” Future collaborations between the companies are expected to focus on peptides for metabolic conditions, Lactocore’s other therapeutic area.
More from Science & Enterprise:
We designed Science & Enterprise for busy readers including investors, researchers, entrepreneurs, and students. Except for a narrow cookies and privacy strip for first-time visitors, we have no pop-ups blocking the entire page, nor distracting animated GIF graphics. If you want to subscribe for daily email alerts, you can do that here, or find the link in the upper left-hand corner of the desktop page. The site is free, with no paywall. But, of course, donations are gratefully accepted.
* * *
|
Welcome to Science & Enterprise Science and Enterprise is an online news service begun in 2010, created for researchers and business people interested in taking scientific knowledge to the marketplace.
On the site’s posts published six days a week, you find research discoveries destined to become new products and services, as well as news about finance, intellectual property, regulations, and employment.
|










 RSS - Posts
RSS - Posts
You must be logged in to post a comment.